Which project are you working on at the moment?
I am comparing the biological process of two most well studied symbionts: arbuscular mycorrhizal fungi and rhizobial bacteria. Plants have evolved to form alliances with microorganisms in the rhizosphere to acquire nutrients that limit their growth. The arbuscular mycorrhizal symbiosis appeared in the very early stages of plant evolution-about 450 mya-while evidence at the rhizobial symbiosis evolving from arbuscular mycorrhizal associations between 60-80 mya. The two share many biological aspects in common: capturing nutrients and trading them for carbon with the plant, secreting specific molecules to be recognized, and entering the plant root to directly associate with the plant cells. However there are also key differences: the formation of nodules that form to harbor rhizobia, which are absent in arbuscular mycorrhizal associations. Moreover, arbuscular mycorrhizal association is prevalent in land plants including cereals whereas rhizobial symbiosis is restricted to a few number of plant species mostly in the legume family. Therefore, to engineer nitrogen fixation in cereals, understanding the differences and the commonalities between these two symbiosis is vital since the arbuscular symbiosis pathway can be piggybacked to form rhizobial symbiosis. Which genes do the plants use in common for both symbiosis? Which genes are pushing them through two different symbiotic processes? How do these genes behave in different contexts such as mutants, time-points and cells? These are my prime questions.
To answer these questions, I am generating and analyzing omics data, mostly transcriptomics, in many mutants and time-points of model legume Medicago truncatula going through the symbiosis program.
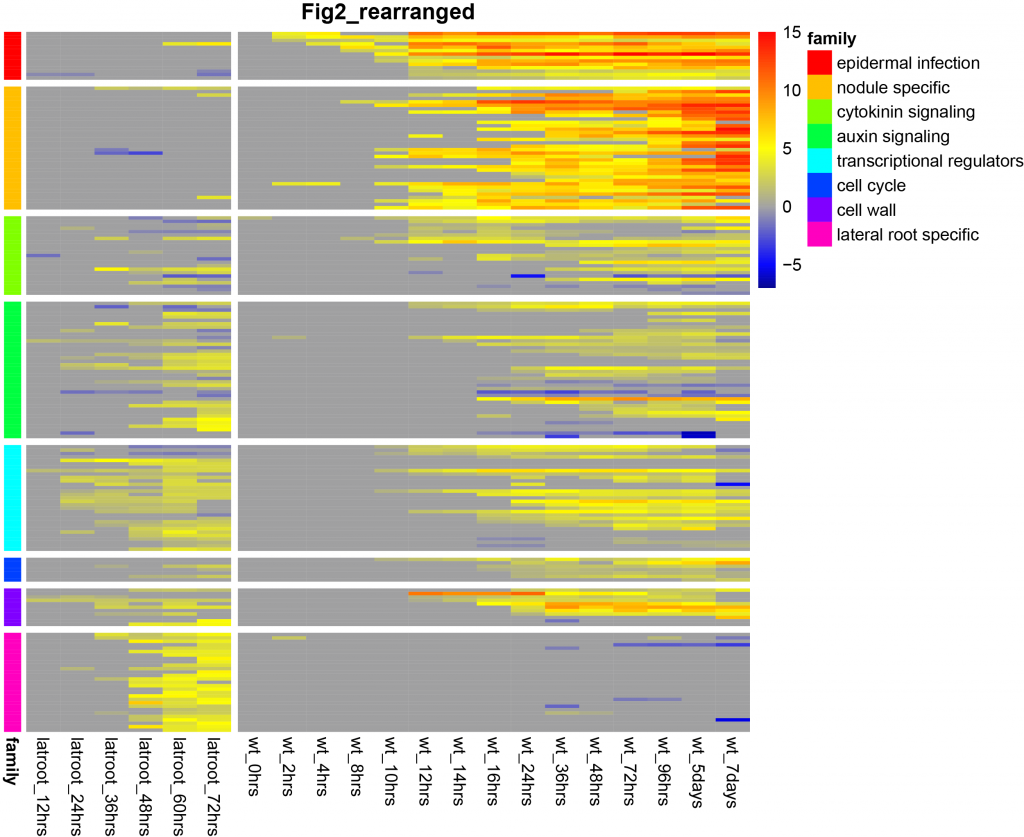
Transcriptomics data comparing the lateral root program to nodule organogenesis program — another project that Tak has contributed to in the ENSA portfolio.
What is a typical day like for you?
Since I am also a bioinformatician, I have to allocate a considerable amount of my time in data analysis and in-silico experiments. So I structure myself to effectively interchange from wet-lab to dry-lab and vice versa. I give my priority to wet-lab experiments since the plant materials would not wait for me. I fill in the dry-lab analysis in between. However, when I go through one full wet-lab experiment and obtain my valuable results, then I invest most of my time in dry-lab data analysis. Most of the time I get something interesting and wet-lab validation is required. Then my cycle starts from wet-lab again. In short, I spend my days interchanging between wet-lab and dry-lab.
What are the hardest parts related to your research?
As I mentioned above, I was trained to be a bioinformatician throughout my Ph.D. I had almost zero experience in wet-lab experiments so it was difficult to shape my mindset to an experimental biologist and to learn the techniques. Interchanging between them was even more difficult for me. However, I had very nice and talented colleagues with me. I’ve learned a lot from them since I’ve started ENSA, and now things are much better for me now.
Why did you choose to work in plant science research?
My grandparents lived in a rural area of South Korea. When I was young, I would visit their home often and enjoyed walking in the woods to observe various plant species. My mother was an expert in distinguishing them. Whenever I walked with her in the woods, she would tell me their names and explain the most that she knew of. I smelled, touched and ate the plants. I got more interested, and wanted to know more about them. Then one day I came across the book ‘The private life of plants’ written by David Attenborough. I got immersed in it, especially in the chapter about organisms that form alliances with the plants. I was fascinated by the relationships between ants-acacia, mycorrhizae-land plants, and algae-fungi which were all benefiting each other. Again I wanted to know more, and yearned to discover the secret of plants myself which led me to be a plant scientist.
Besides your scientific interests, what are your personal interests?
I enjoy playing table tennis in my free time. It is a sport that is easy to learn but difficult to master. Very subtle movements of your body can lead to big changes in the course and the spin of the ball. This makes it more attractive to me since I find it challenging and needs much engagement so I’ll never get bored of it. It is also a very competitive sport because most of the time it is a 1-to-1 match with an opponent and winning or losing is solely your responsibility. Victory is more rewarding but defeat is so much bitter than other sports. So I play my utmost for the sweet juice of triumph and I enjoy it very much to defeat my opponents.
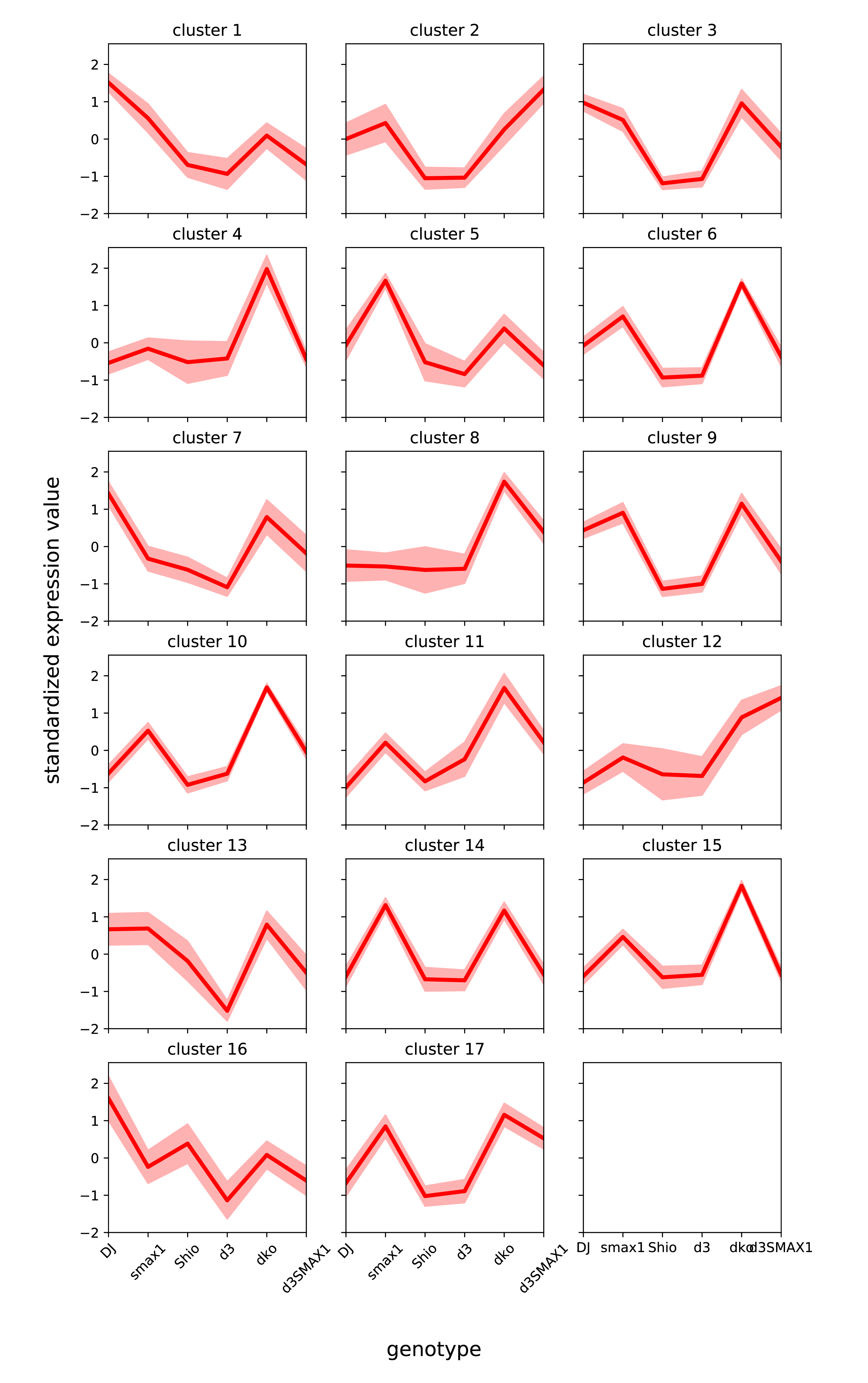
Identifying gene patterns in rice mutants that is related to mycorrhizal symbiosis (copyright Tak Lee).

